Note
Click here to download the full example code
Plotting validation curves¶
In this example the impact of the Geometric SMOTE’s hyperparameters is examined. The validation scores of a Geometric SMOTE-GBC classifier is presented for different values of the Geometric SMOTE’s hyperparameters.
# Author: Georgios Douzas <gdouzas@icloud.com>
# Licence: MIT
import matplotlib.pyplot as plt
import numpy as np
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import LinearSVC
from sklearn.model_selection import validation_curve
from sklearn.metrics import make_scorer, cohen_kappa_score
from sklearn.datasets import make_classification
from imblearn.pipeline import make_pipeline
from imblearn.metrics import geometric_mean_score
from gsmote import GeometricSMOTE
print(__doc__)
RANDOM_STATE = 10
SCORER = make_scorer(geometric_mean_score)
def generate_imbalanced_data(weights, n_samples, n_features, n_informative):
"""Generate imbalanced data."""
X, y = make_classification(
n_classes=2,
class_sep=2,
weights=weights,
n_informative=n_informative,
n_redundant=1,
flip_y=0,
n_features=n_features,
n_clusters_per_class=2,
n_samples=n_samples,
random_state=RANDOM_STATE,
)
return X, y
def generate_validation_curve_info(estimator, X, y, param_range, param_name, scoring):
"""Generate information for the validation curve."""
_, test_scores = validation_curve(
estimator,
X,
y,
param_name=param_name,
param_range=param_range,
cv=3,
scoring=scoring,
n_jobs=-1,
)
test_scores_mean = np.mean(test_scores, axis=1)
test_scores_std = np.std(test_scores, axis=1)
return test_scores_mean, test_scores_std, param_range
def plot_validation_curve(validation_curve_info, scoring_name, title):
"""Plot the validation curve."""
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
test_scores_mean, test_scores_std, param_range = validation_curve_info
plt.plot(param_range, test_scores_mean)
ax.fill_between(
param_range,
test_scores_mean + test_scores_std,
test_scores_mean - test_scores_std,
alpha=0.2,
)
idx_max = np.argmax(test_scores_mean)
plt.scatter(param_range[idx_max], test_scores_mean[idx_max])
plt.title(title)
plt.ylabel(scoring_name)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.get_xaxis().tick_bottom()
ax.get_yaxis().tick_left()
ax.spines['left'].set_position(('outward', 10))
ax.spines['bottom'].set_position(('outward', 10))
plt.ylim([0.9, 1.0])
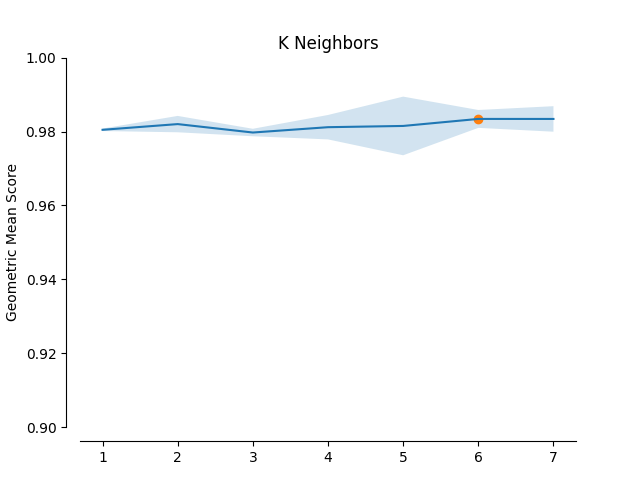
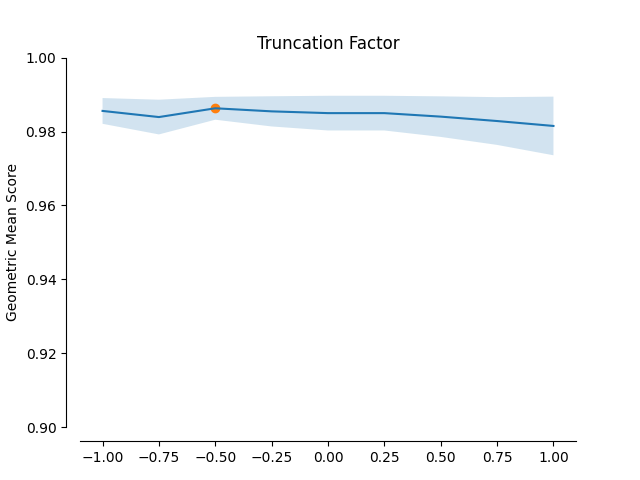
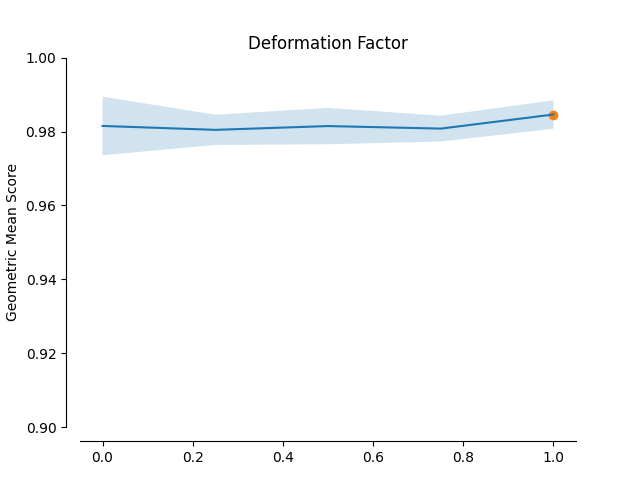
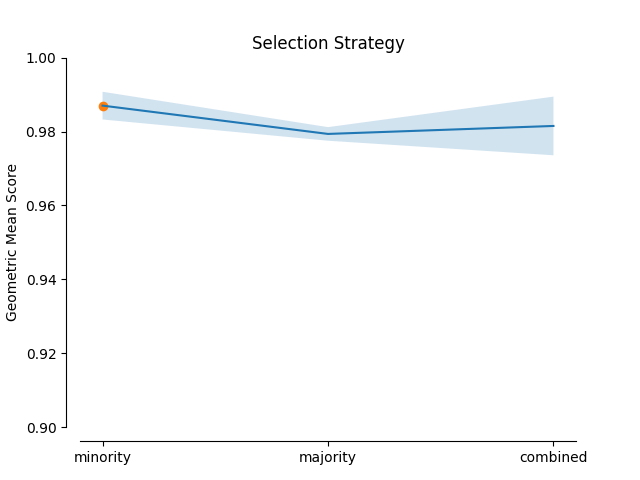
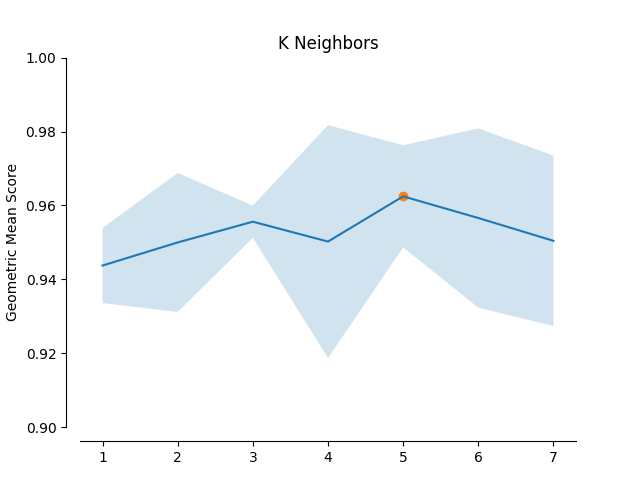
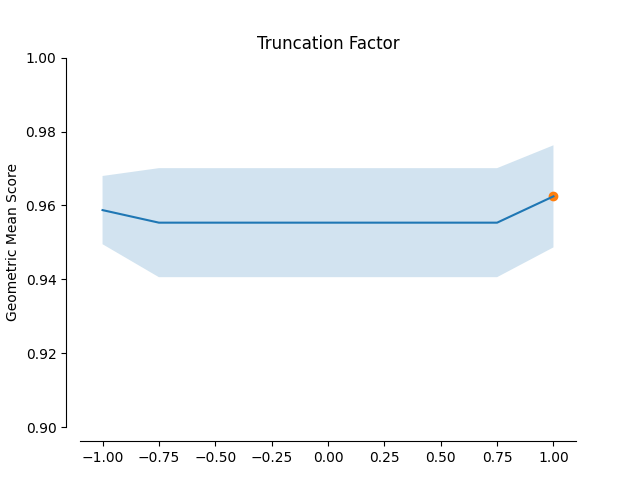
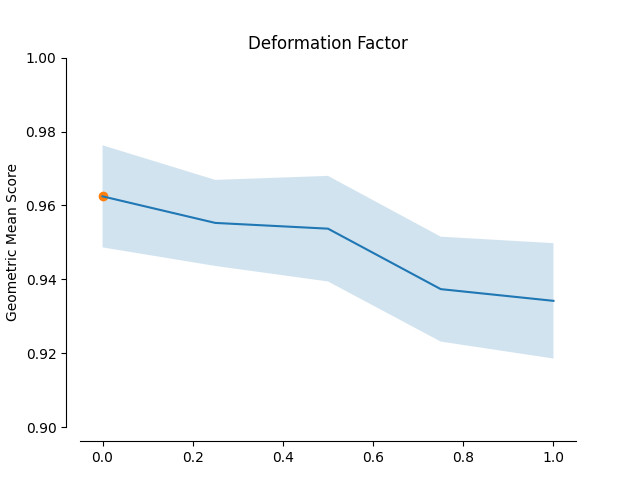
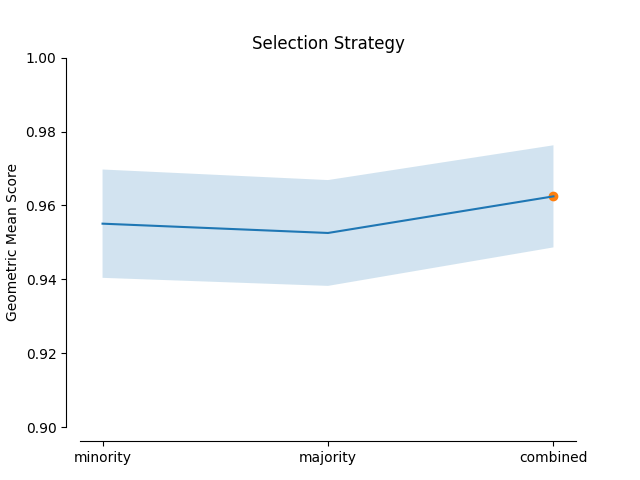
Low Imbalance Ratio or high Samples to Features Ratio¶
When  (Imbalance Ratio) is low or
(Imbalance Ratio) is low or 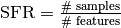 (Samples to Features Ratio) is high then the
minority selection strategy and higher absolute values of the truncation and
deformation factors dominate as optimal hyperparameters.
(Samples to Features Ratio) is high then the
minority selection strategy and higher absolute values of the truncation and
deformation factors dominate as optimal hyperparameters.
X, y = generate_imbalanced_data([0.3, 0.7], 2000, 6, 4)
gsmote_gbc = make_pipeline(
GeometricSMOTE(random_state=RANDOM_STATE),
DecisionTreeClassifier(random_state=RANDOM_STATE),
)
scoring_name = 'Geometric Mean Score'
validation_curve_info = generate_validation_curve_info(
gsmote_gbc, X, y, range(1, 8), "geometricsmote__k_neighbors", SCORER
)
plot_validation_curve(validation_curve_info, scoring_name, 'K Neighbors')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
np.linspace(-1.0, 1.0, 9),
"geometricsmote__truncation_factor",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Truncation Factor')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
np.linspace(0.0, 1.0, 5),
"geometricsmote__deformation_factor",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Deformation Factor')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
['minority', 'majority', 'combined'],
"geometricsmote__selection_strategy",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Selection Strategy')
High Imbalance Ratio or low Samples to Features Ratio¶
When 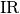 is high or
is high or 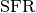 is low then the majority
or combined selection strategies and lower absolute values of the truncation
and deformation factors dominate as optimal hyperparameters.
is low then the majority
or combined selection strategies and lower absolute values of the truncation
and deformation factors dominate as optimal hyperparameters.
X, y = generate_imbalanced_data([0.1, 0.9], 2000, 400, 200)
gsmote_gbc = make_pipeline(
GeometricSMOTE(random_state=RANDOM_STATE),
LinearSVC(random_state=RANDOM_STATE, max_iter=1e5),
)
scoring_name = 'Geometric Mean Score'
validation_curve_info = generate_validation_curve_info(
gsmote_gbc, X, y, range(1, 8), "geometricsmote__k_neighbors", SCORER
)
plot_validation_curve(validation_curve_info, scoring_name, 'K Neighbors')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
np.linspace(-1.0, 1.0, 9),
"geometricsmote__truncation_factor",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Truncation Factor')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
np.linspace(0.0, 1.0, 5),
"geometricsmote__deformation_factor",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Deformation Factor')
validation_curve_info = generate_validation_curve_info(
gsmote_gbc,
X,
y,
['minority', 'majority', 'combined'],
"geometricsmote__selection_strategy",
SCORER,
)
plot_validation_curve(validation_curve_info, scoring_name, 'Selection Strategy')
Total running time of the script: ( 0 minutes 35.286 seconds)